
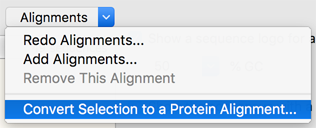
If a feature is interrupted by insertion of another sequence, its coordinates can be defined as two disjoint regions using the Split Feature option (#4 in Figure 1, see example in Figure 2).įigure 3.
If the feature is an ORF, “N/A” should be entered in the /product field. For example, if the merA gene is interrupted, two features- merA 5’-end and merA 3’-end-should be created.
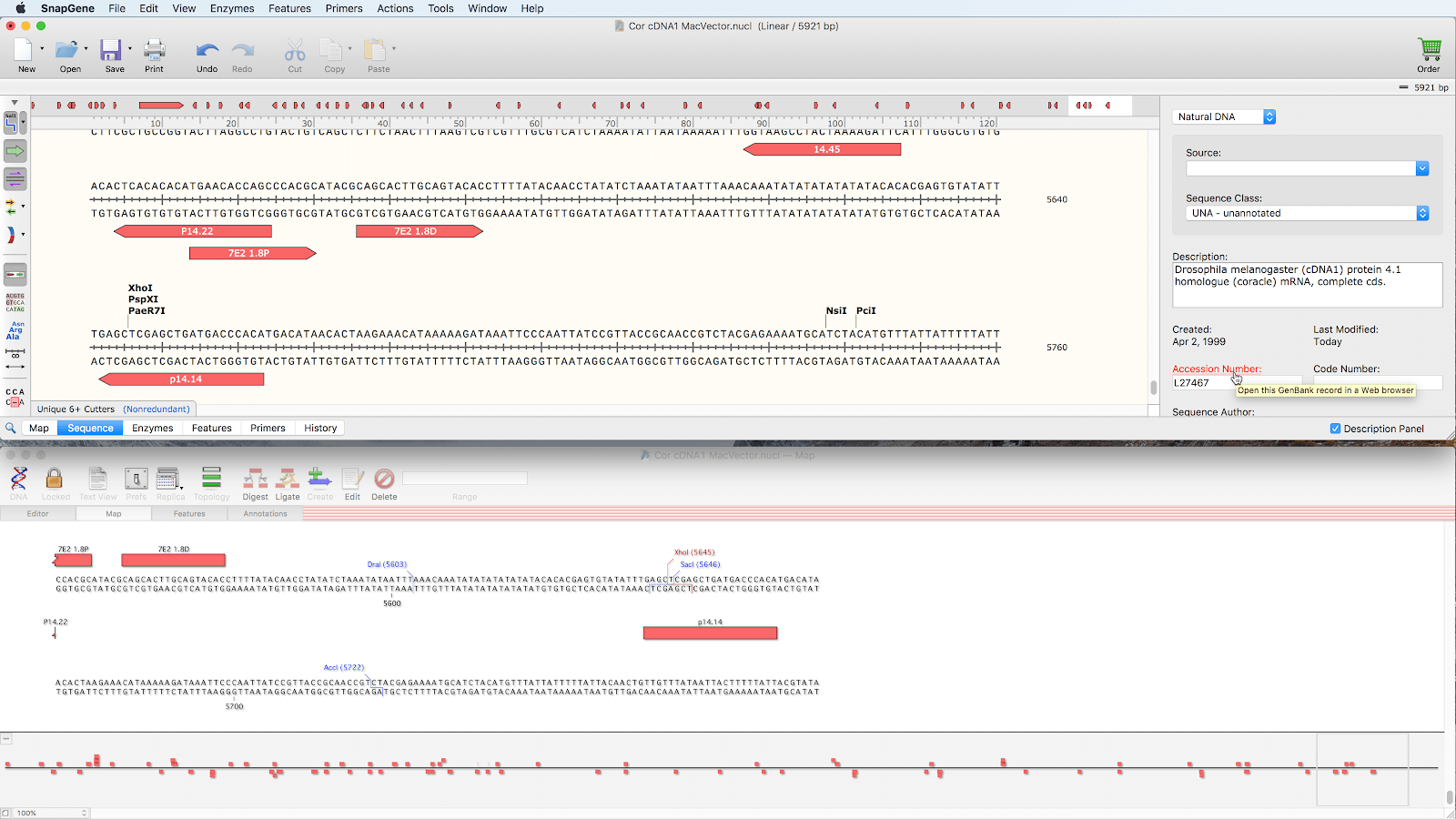
Name (#2 in Figure 1) is a sub-field within the /note field for some feature types. Label (#1 in Figure 1) refers to the text entered in the Feature text box in the SnapGene feature editing interface.Multiple items within any annotation field should be separated by “ ||”.Therefore, for each feature which we do want to extract, we include a field in /Note Capture = yes. In some cases, the Genbank files have additional features that we are not interested in capturing for TnCentral.These features will all be documented in the enhanced Genbank files according to the guidelines below. 10.3 Repeat Regions and Recombination Sitesįor TnCentral, we are interested in curating the following features: Protein-coding genes (e.g., transposase genes, accessory genes, passenger genes), Mobile Elements (e.g., transposons, insertion sequences, integrons), Repeat Elements (e.g., IRL, IRR), and Recombination Sites (e.g, Res and attC).10 Field Structure for /note for Different Feature Types.9.5 Annotation of Recombination Sites in Mobile Element Sequence Variants.9.2 Feature Type (selected from SnapGene Pull-Down Menu).8.5 Annotation of Repeat Elements in Mobile Element Sequence Variants.8.2 Feature Type (selected from SnapGene Pull-Down Menu).
7 Annotation of CDS in Mobile Element Sequence Variants.6.6 Passenger Genes: Subclass Hypothetical.6.4 Passenger Genes: Subclass Antibiotic Resistance (ABR) Genes.6.3 Passenger Genes: Subclass Toxin and Antitoxin Genes.6 Further Information About Annotation of Some Specific Types of CDS.5.3 Feature Type (selected from SnapGene Pull-Down Menu).4 Annotation of Internal Mobile Elements.3.3 Mobile Element Type and Name (/mobile_element_type field).3.2.1 Feature Type (selected from SnapGene Pull-Down Menu).
SNAPGENE FEATURE DATABASE DOWNLOAD
2.1 Download our Snapgene curated Library.


 0 kommentar(er)
0 kommentar(er)
